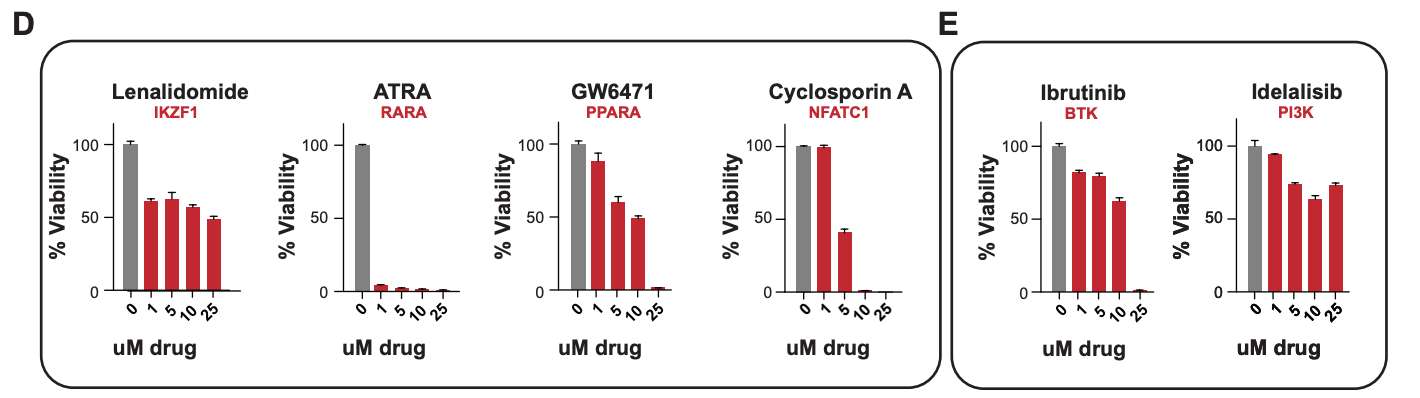
Transcriptional Network Modeling
The control of gene expression in human cells is very poorly understood. My work involves integrating data to design and build models to understand the regulation key genes, especially in cancer. You can download software for one common approach we use here, and read about the method in this preprint. My current focus is integrating high resolution sequencing information called transcription factor footprints to precisely locate the exact base-pairs being bound by regulatory proteins. By applying these methods to cancer, we have identified the cell of origin of obscure cancers and have predicted new sensitivities for drug repurposing.
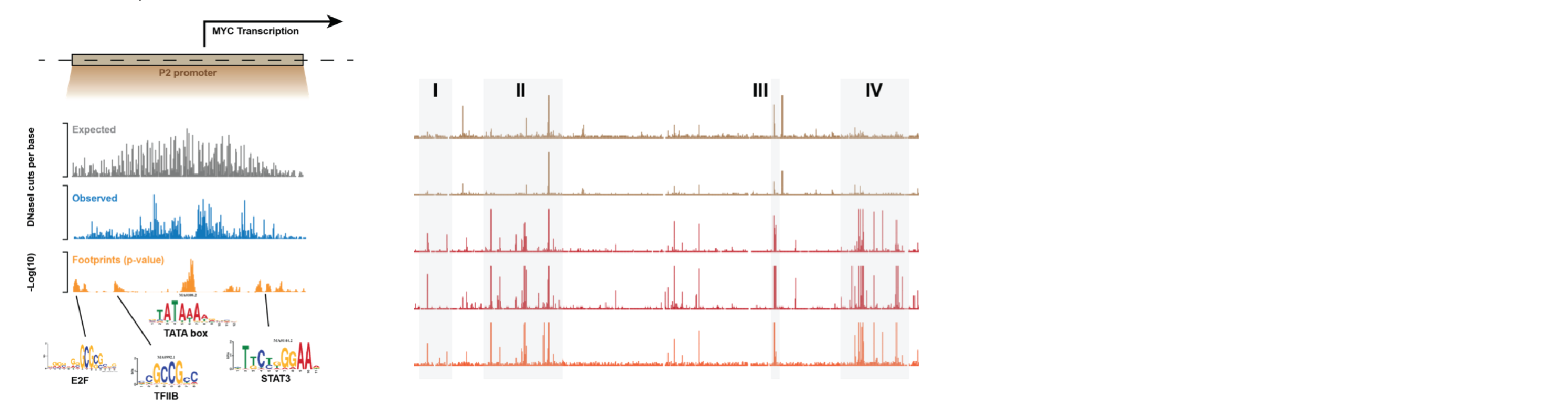
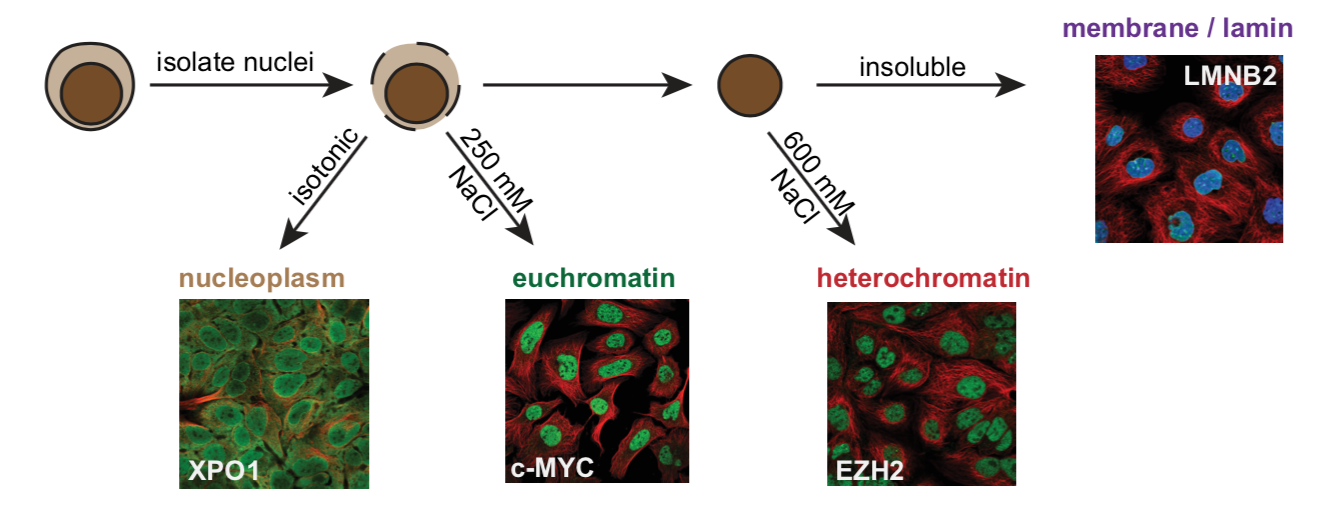
Chromatin Proteomics
In order to build accurate network models, we can’t just rely on sequence-based information, due to the fact that transcription factors within a family bind highly similar operator sequences. A better approach is to directly quantify the proteins bound to chromatin in a cell type of interest. Good methods for this didn’t previously exist, so we developed Chromatin Extraction by Salt Separation followed by Data-Independent Acquisition (ChESS-DIA). With this, we can measure the abundance of proteins freely diffusing in the nucleus, bound in euchromatin and in heterochromatin. See the preprint describing the method.
Chemical Biology
Transcription happens on a relatively fast time-scale, faster than we can do genetics. Therefore, to perturb components of the system and see the direct outcome of the perturbation, we need chemical tools that work quickly and specifically for a target of interest. My focus in chemical biology has been on members of the polycomb repressive complexes, the BET bromodomains and the YEATS chromatin-binding proteins.